NUCLEIC ACIDS
- Nucleic acids are the polymers of nucleotide made up of carbon, hydrogen, oxygen, nitrogen and phosphorus which control the basic functions of the cell.
- Miescher discovered nucleic acids in the nucleus of pus cell and called it nuclein. The name nucleic acid was proposed by Altman.
- On the basis of structure, nitrogen bases are broadly of two types :
- Pyrimidines : These consist of one pyrimidine ring. Skeleton of ring is composed of two nitrogen and four carbon atoms. E.g., cytosine, thymine (in DNA) and uracil (in RNA).
- Purines : These consist of two rings i.e. one pyrimidine ring (2N + 4C) and one imidazole ring (2N + 3C). E.g., adenine and guanine.
PENTOSE SUGAR
- Nitrogen base forms bond with first carbon of pentose sugar to form a nucleoside, nitrogen of third place (N3) forms bond with sugar in case of pyrimidines while in purines nitrogen of ninth place (N9) forms bond with sugar.
- Phosphate : Phosphate forms ester bond (covalent bond) with fifth carbon of sugar to form a complete nucleotide.
- When nitrogen bases are found attached to a pentose sugar, then they are called nucleosides, e.g., adenosine, guanosine, thymidine, uridine and cytidine.
- Nucleotides are phosphorylated nucleosides. These are formed by condensation of a pentose sugar, a nitrogen base and at least one phosphoric acid residue, e.g., adenylic acid, thymidylic acid, guanylic acid, uridylic acid and cytidylic acid.
- Nucleic acids like DNA and RNA consist of nucleotides only. DNA and RNA function as genetic material.
DNA (DEOXYRIBONUCLEIC ACID)
- DNA was first identified and isolated by Friedrich Miescher. A nucleic acid containing deoxyribose is called deoxyribonucleic acid (DNA).
- In DNA, pentose sugar is deoxyribose sugar and four types of nitrogen bases are Adenine (A), Thymine (T), Guanine (G) and Cytosine (C).
- Wilkins and Franklin studied DNA molecule with the help of X-ray crystallography.
- With the help of this study, Watson and Crick (1953) proposed a double helix model for DNA. For this model, Watson, Crick and Wilkins were awarded noble prize in 1962.
- According to this model, DNA is composed of two polynucleotide chains. Both polynucleotide chains are complementary and anti-parallel to each other.
- In both strands of DNA, direction of phosphodiester bond is opposite, i.e., if direction of phosphodiester bond in one strand is 3'-5' then it is 5'-3' in another strand.
- Both strands of DNA are held together by hydrogen bonds. These hydrogen bonds are present between nitrogen bases of both strands.
- Adenine binds to Thymine by two hydrogen bonds and Cytosine binds to Guanine by three hydrogen bonds.
- Chargaff’s equivalence rule : In a double stranded DNA, amount of purine nucleotides is equal to amount of pyrimidine nucleotides.
Purine = Pyrimidine
[A] + [G] = [T] + [C]
Base ratio = 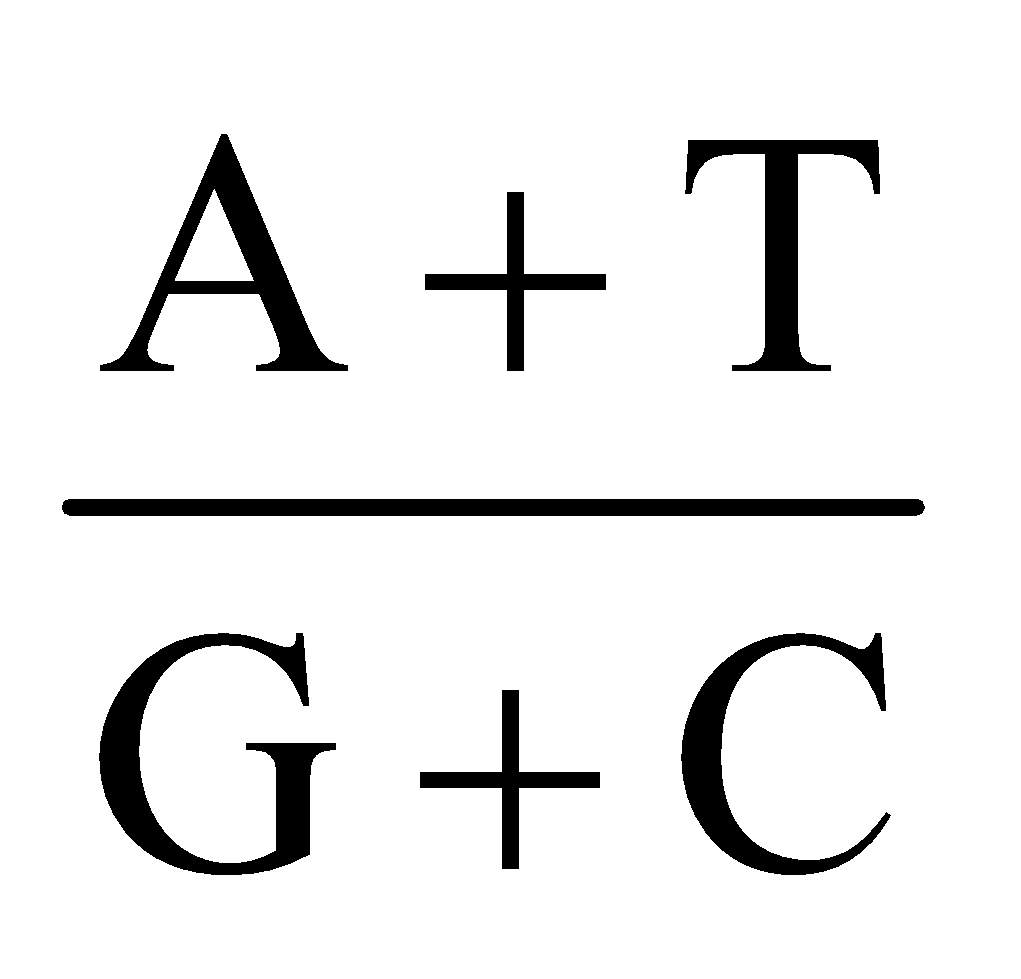 = constant for a given species.
= constant for a given species.
In DNA , A + T > G + C ⇒ A – T type DNA.
Base ratio of A – T type of DNA is more than one.
E.g., Eukaryotic DNA.
In DNA, G + C > A + T ⇒ G – C type DNA.
Base ratio of G - C type of DNA is less than one.
E.g., Prokaryotic DNA.
- Melting point of DNA depends on G – C contents.
More G – C content means more melting point.
Tm (melting temperature) of prokaryotic DNA > Tm of eukaryotic DNA.
CONFIGURATION OF DNA MOLECULE
- Two strands of DNA are helically coiled like a revolving ladder. Backbone of this ladder (Reiling) is composed of phosphates and sugars while steps (bars) are composed of pairs of nitrogen bases. The strand turns 36°.
- Distance between two successive steps is 3.4 Å, In one complete turn of DNA molecule, there are 10 such steps
(10 pairs of nitrogen bases). So, the length of one complete turn is 34 Å. This is called helix length. - Diameter of DNA molecule i.e., distance between phosphates of two strands is 20Å.
- Distance between sugar of two strands is 11.1Å.
- Length of hydrogen bonds between nitrogen bases is 2.8-3.0Å. Angle between nitrogen base and C1 carbon of pentose is 51°.
- Molecular weight of DNA is 106 to 109 dalton.
- In the nucleus of eukaryotes, the DNA is associated with histone proteins to form nucleoprotein.
- Bond between DNA and histone is salt linkage (Mg+2).
- DNA in chromosomes is linear while in prokaryotes, mitochondria and chloroplast, it is circular.
- In φ × 174 bacteriophage, the DNA is single stranded and circular.
- G-4, S-13, M-13, F1 and Fd - Bacteriophages also contain ss (single stranded) - circular DNA.
TYPES OF DNA
On the basis of direction of twisting, there are two types of DNA:
RIGHT HANDED DNA
- Clockwise twisting e.g. The DNA for which Watson and Crick proposed model was B - DNA.
- Other exmples of right handed DNA are –
LEFT HANDED DNA
- Anticlockwise twisting e.g. Z-DNA-discovered by Alexander Rich.
- Phosphate and sugar backbone is zig-zag.
- Units of Z-DNA are dinucleotides (purine and pyrimidine in alternate order)
- Helix length 45.6 Å, Diameter - 18.4 Å
- No. of base pairs : 12 (6 dimers)
- Distance between base pairs = 3.75 Å
FUNCTIONS OF DNA
DNA is vital for all living beings – It is important for inheritance, coding for proteins and the genetic instruction guide for life and its processes. DNA holds the instructions for an organisms or each cell's development and reproduction and ultimately death.
NOTES
- DNA molecule is Dextrorotatory while RNA molecule is Laevorotatory.
- C-value = Total amount of DNA in a genome or haploid set of chromosomes.
RNA (RIBONUCLEIC ACID)
Structure of RNA is fundamentally the same as DNA, but there are some differences. The differences are :
- In place of deoxyribose sugar of DNA, ribose sugar is present in RNA.
- In place of nitrogen base thymine present in DNA, nitrogen base uracil is present in RNA.
- RNA is made up of only one polynucleotide chain i.e., RNA is single stranded.
Exception : RNA found in reo-virus is double stranded, i.e., it has two polynucleotide chains.
TYPES OF RNA
GENETIC RNA OR GENOMIC RNA
In the absence of DNA, sometimes RNA working as genetic material and genomic RNA transfer information from one generation to the next generation.
E.g., Reovirus and Tobacco Mosaic Virus (TMV).
NON-GENETIC RNA
3 types : (a) r - RNA (b) t - RNA (c) m - RNA
(1) Ribosomal RNA (r - RNA)
- This RNA is 80% of the cells total RNA.
- It is found in ribosomes and is produced in the nucleolus.
- At the time of protein synthesis, r-RNA provides attachment sites to t-RNA and m-RNA and attaches them on the ribosome.
(2) Transfer - RNA (t-RNA) / soluble RNA (sRNA) / adapter RNA
- It is 10-15% of total RNA.
- It is synthesized in the nucleus by DNA.
(3) Messenger RNA (m -RNA)
- The m- RNA is 1-5% of the cell’s total RNA.
- It is produced by genetic DNA in the nucleus. This process is known as transcription.
DYNAMIC STATE OF BODY CONSTITUENTS - CONCEPT OF METABOLISM
- Metabolism is the set of life sustaining chemical transformations within the cells of living organisms. These enzyme - catalyzed reactions allows organisms to grow, reproduce, maintain their structures, and respond to their environments.
- A few examples for such metabolic transformations are : removal of CO2 from amino acids making an amino acid into an amine, removal of amino group in a nucleotide base; hydrolysis of a glycosidic bond in a disaccharide, etc.
- It is divided into 2 classes : Catabolism breaks down organic matter, e.g., to harvest energy in cellular respiration. Anabolism uses energy to construct components of cells such as proteins and nucleic acids.
- Metabolites are converted into each other in a series of linked reactions by a sequence of enzymes called metabolic pathways.
- In metabolic reactions, every chemical reaction is catalysed reaction. There is no uncatalysed metabolic conversion in living systems.
- Metabolic Basis for Living
- Metabolic pathways can lead to a more complex structure from a simpler structure (for example, acetic acid becomes cholesterol) or to a simpler structure from a complex structure (for example, glucose becomes lactic acid in our skeletal muscles).
- Living organisms have learnt to trap energy liberated during degradation and store it in the form of chemical bonds. As and when needed, this bond energy is utilised for biosynthetic, osmotic and mechanical work that we perform. The most important form of energy currency in living systems is the bond energy in a chemical called adenosine triphosphate (ATP).
ENZYMES
- Enzymes are biocatalysts made up of proteins (except ribozyme) which increases the rate of biochemical reactions by lowering down the activation energy, but does not affect the nature of final product.
- The term enzyme (meaning in yeast) was used by Willy Kuhne (1878) while working on fermentation.
- Zymase (from yeast) was the first discovered enzyme by Buchner.
- The first purified and crystalized enzyme was urease
(by J.B. Sumner) from Canavalia/Jack Bean (Lobia plant) and suggested that enzymes are proteins.
CHARACTERISTICS OF ENZYMES
- All enzymes are proteins, but all proteins are not enzymes. Enzymatic proteins consist of 20 amino acids.
- All enzymes are tertiary and globular proteins (isoenzymes quaternary protein). Their tertiary structure is very specific and important for their biological activity.
- Enzymes accelerate the rate of reaction without undergoing any change in themselves. Enzymes lower the activation energy of substrate or reactions.
- Enzymes are macromolecules of amino acids which are synthesized on ribosomes under the control of genes.
- Molecular weight of enzymes are high and these are colloidal substances.
- Enzymes are very sensitive to pH and temperature. Optimum temperature for enzymes is 20-35°C. Most of the enzymes are active at neutral pH, hydrolytic enzymes of lysosomes are active at acidic pH (5).
- Enzymes are required in very minute amounts for biochemical reactions.
- Their catalytic power is represented by Michaelis Menten constant or Km constant and turn over number. ‘‘The number of substrate molecules converted into products per unit time by one molecule of the enzyme in favourable conditions is called turnover number.’’
- Enzymes are very specific to their substrate or reactions.
STRUCTURE OF ENZYMES
- Simple enzymes : These are exclusively made up of protein i.e., simple proteins.
E.g., pepsin, trypsin, papain.
- Conjugated enzymes : Enzymes are composed of one or several polypeptide chains. However, there are a number of cases in which non-protein constituents called co-factors are bound to the enzyme to make the enzyme catalytically active. The protein part of the enzyme is called apoenzyme. There are 3 kinds of cofactors which are given as follows :
- Co-enzymes : These are non-protein organic groups, which are loosely attached to apoenzymes. These are generally made up of vitamins, e.g., coenzyme nicotinamide adenine dinucleotide (NAD), nicotinamide adenine dinucleotide phosphate (NADP) contains the vitamin niacin, coenzyme A contains pantothenic acid, flavin mononucleotide (FMN), flavin adenine dinucleotide (FAD) contains riboflavin (Vitamin B2), and thiamine pyrophosphate (TPP) contains thiamine (Vitamin B1).
- Prosthetic group : When non-protein part is tightly or firmly attached to apoenzyme. These are organic compounds. E.g., in peroxidase and catalase, which catalyze the breakdown of H2O2 to H2O and O.
- Metal ions : Metal ions play an essential role in regulating the activity of enzymes by forming coordination bonds with side chains at the active site and at the same time form one or more coordination bonds with the substrate. E.g., Mn, Fe, Co, Zn, Ca, Mg, Cu.
- Active site : Specific part of enzyme at which specific substrate is to bind and catalyse the reaction is known as an active site. Active site of enzyme is made up of very specific sequence of amino acids which is, determined by genetic codes.
- Allosteric site : Besides the active site, some enzymes possess additional sites, at which chemicals other than substrate (allosteric modulators) bind. These sites are known as allosteric sites and enzymes with allosteric sites are called as allosteric enzymes, e.g., hexokinase, phosphofructokinase.
TERMS RELATED TO ENZYMES
- Endoenzymes : Enzymes which are functional only inside the cells.
- Exoenzymes : Enzymes catalysed the reactions outside the cell. E.g., enzymes of digestion, some enzymes of insectivorous plants, zymase complex of fermentation.
- Proenzyme/Zymogen : These are precursors of enzymes or inactive forms of enzymes.
E.g., Pepsinogen, Trypsinogen, etc.
- Iso-enzymes : Enzymes having similar action, but little difference in their molecular configuration are called isoenzymes. 16 forms of α-amylase of wheat and 5 forms of LDH (Lactate dehydrogenase) are known. These all forms are synthesised by different genes.
- Inducible enzymes : When formation of enzyme is induced by substrate availability. E.g., Lactase, Nitrogenase, β-galactosidase.
- Biodetergents : Enzymes used in washing powders are known as bio-detergents, e.g., amylase, lipase, proteolytic enzymes.
- Housekeeping / constitutive enzymes : These enzymes are always present in constant amount and are also essential to cell.
NOMENCLATURE AND CLASSIFICATION OF ENZYMES
MECHANISM OF ENZYME ACTION
- Energy is required to convert the inert molecules into the activated state. The amount of energy required to raise the energy of molecules at which chemical reaction can occur is called activation energy.
- Enzymes act by decreasing the activation energy so that the number of activated molecules is increased at lower energy levels. If the activation energy required for the formation of the enzyme-substrate complex is low, many more molecules can participate in the reaction than would be the case if the enzymes were absent.
MODE OF ACTION OF ENZYME
(1) LOCK & KEY THEORY OR TEMPLATE THEORY
- The theory was given by Emil Fischer.
- According to this theory, active sites of enzymes serve as a lock into which the reactant substrate fits like a key. Enzymes have specific sites where a particular substrate can only be attached. This model accounts for enzyme specificity.
(2) ENZYME - SUBSTRATE COMPLEX THEORY
In 1913, Michaelis and Menten proposed that for a catalytic reaction to occur it is necessary that the enzyme and substrate bind together to form an enzyme substrate complex.
It is amazing that the enzyme-substrate complex breaks up into chemical products different from that which participated in its formation (i.e., substrates). On the surface of each enzyme, there are many specific sites for binding substrate molecules called active sites or catalytic sites.
(3) INDUCED FIT THEORY
- This hypothesis was proposed by Koshland (1959).
- According to this theory, active site is not static but it undergoes a conformational change which is induced by specific substrate.
FACTORS / AFFECTING ENZYME ACTION
The activity of an enzyme can be affected by a change in the conditions which can alter the tertiary structure of the protein. These include temperature, pH, change in substrate concentration or binding of specific chemicals that regulate its activity.
- pH : Enzymes are very sensitive to pH. Each enzyme shows its highest activity at optimum pH. Activity declines both below and above the optimum value.
- Temperature : Low temperature preserves the enzyme in a temporarily inactive state whereas high temperature destroys enzymatic activity because proteins are denatured by heat. Generally all enzymes perform better at body temperature of an organism.
- Enzyme concentration : The rate of reaction is directly proportional to enzyme concentration. An increase in enzyme concentration will cause a rise in the rate of reaction upto a point and after which the rate of reaction becomes constant. Increasing the enzyme concentration, increases the number of available active sites.
- Substrate concentration : Increase in substrate concentration increases the activity of enzymes until all the active sites of enzymes are saturated by the substrate molecules. Therefore, the substrate molecules occupy the active sites vacated by the products and cannot increase the rate of reaction further.
Km CONSTANT (MICHAELIS & MENTEN CONSTANT)
- Km constant of an enzyme is the concentration of substrate at which rate of reaction of that enzyme attains half of its maximum velocity. It is given by Michaelis & Menten. The value of Km should be lower for an enzyme.
- Km exhibits catalytic activity of an enzyme.
- Km value differs from substrate to substrate because different enzymes differ in their affinity towards different substrates. A high Km indicates low affinity while a low Km shows strong affinity.
- Protease acts on different proteins. So its Km value differs from protein to protein.
The Michaelis Menten equation describes how rate of reaction relatively varies with substrate concentration
V0 = 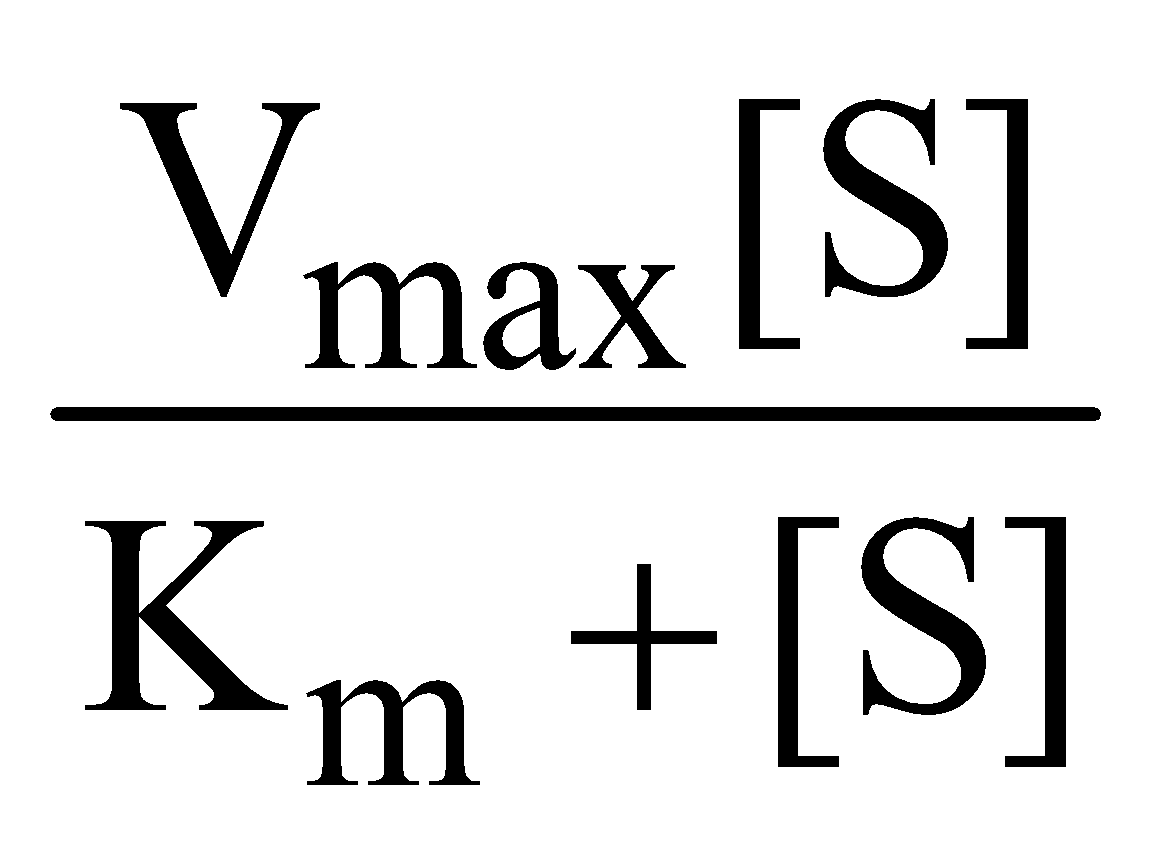
Where Vo is the rate of initial reaction; Vmax is the maximum relative or the reaction rate with excess substrate; Km is the Michaelis constant = K2+K3/K1; [S] is the substrate concentration.
The above reaction shows that the greater the affinity between an enzyme and its substrate, the lower the Km (in units moles per litre) of the enzyme substrate reaction. Stated inversely, 1/Km is the measure of affinity of the enzyme for its substrate.
